- Bootstraps of the likelihood ratio, holding the parameters fixed, finally completed.
Anoles: 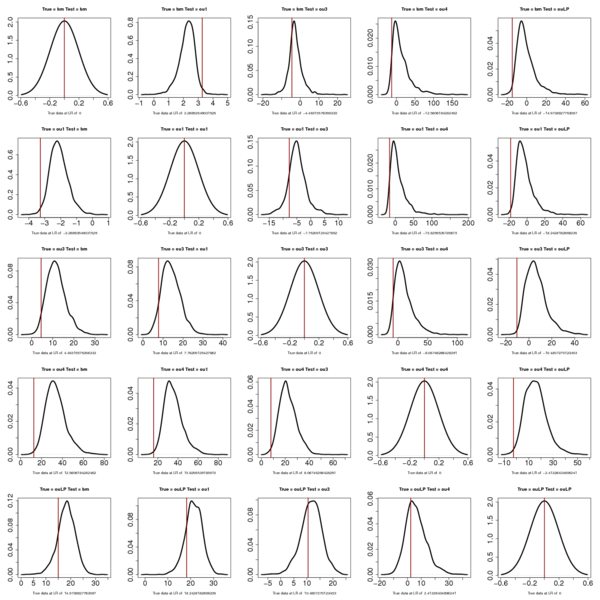
Summary Anoles table:
> summary(anoles_boots)
[,1] [,2] [,3] [,4] [,5]
[1,] 0.500000000 0.981399498 0.26514573 0.06668672 0.018154938
[2,] 0.009580545 0.500000000 0.15610274 0.04819938 0.011616566
[3,] 0.039233893 0.075349644 0.50000000 0.05699877 0.005443493
[4,] 0.004211230 0.004592917 0.01389950 0.50000000 0.006648916
[5,] 0.173457542 0.198303402 0.38893099 0.33891430 0.500000000
>Labrids: 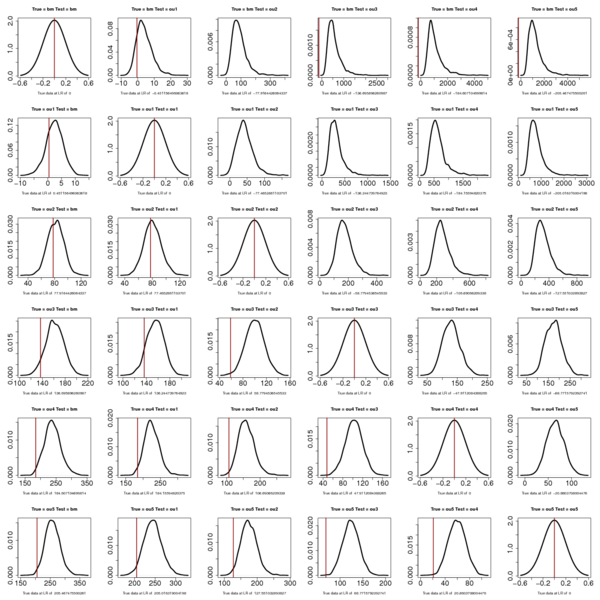
> summary(labrid_boots)
[,1] [,2] [,3] [,4]
[1,] 0.5000000 6.036842e-05 0.0000000 0.0
[2,] 0.5424613 5.000000e-01 0.0000000 0.0
[3,] 0.5546890 4.384114e-01 0.5000000 0.0
[4,] 0.4081957 3.488395e-01 0.0295128 0.5
>
Note that the location of the red line now just flips sign in the pairwise comparison.
- Functions for bootstrapping of the likelihood directly also written. Currently running with 1000 replicates.
- model_bootstrap() bootstraps a single model
- model_bootstrap_all() applies this to a list of models
- plot.model_boots() plots the object returned by bootstrapping all models on a single plot axis.
- Working on a summary function for both LR and likelihood bootstraps.