Implemented in BUGS/allen.md
- [x] Inverse gamma priors on variances 36888da
- [x] Uniform prior on standard deviations 75df941
- [x] Mean plot for parametric fit 75df941
[x] Convergence diagnostics for both parametric and nonparametric MCMC, using similar visual layout 75df941
Oh, Jeromy Anglim has a rather nice collection of jags links
On Variance Priors for the Parametric MCMC
- Gelman recommends uniform priors on standard deviations for the noise terms: in the
.bugsfile we have
stdQ ~ dunif(0,100)
stdR ~ dunif(0,100)
# as "precision" tau instead of stdev sigma
iQ <- 1/(stdQ*stdQ);
iR <- 1/(stdR*stdR);Where these enter the model as
for(t in 1:(N-1)){
mu[t] <- x[t] + exp(r0 * (1 - x[t]/K)* (x[t] - theta) )
x[t+1] ~ dnorm(mu[t],iQ)
}
for(t in 1:(N)){
y[t] ~ dnorm(x[t],iR)
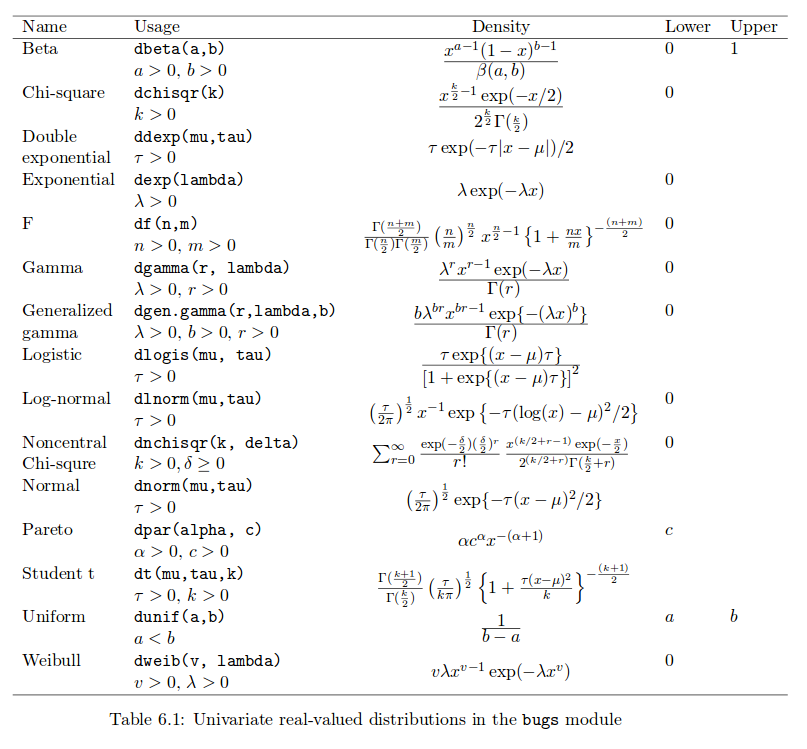
}Note that dnorm in JAGS notation is defined in terms of the mean and the precision (reciprocal of the variance), rather than standard deviation (e.g. in R’s dnorm function), see the JAGS manual, particularly Table 6.1 pg 29, below. Note that the standard deviation has the uniform prior, not the precision.
- Also try inverse gamma priors,
iQ ~ dgamma(0.0001,0.0001)
iR ~ dgamma(0.0001,0.0001)(Precision is Gamma distributed when the variances are inverse-gamma distributed.
- Both are quite uniformative, results appear quite comparable.
Markov Chain Monte Carlo Convergence Analysis
- Add for both parametric and non-parametric cases
- Extra long runs before finalizing analysis
- Some examples with R code
A variety of tools from coda package. Of course these methods are designed from toy problems and cannot guarentee convergence.
- Traceplots, density plots
- Running mean, autocorrelation plots
autocorr.plot - Metropolis acceptance/rejection rate
rejectionRate - Multiple chain diagonostics: Gelman-Rubin
gelman.diag,gelman.plot - Geweke (compare means between different sections)
geweke.diag - Raferty-Lewis
raferty.diag - Heidelberg and Welch
heidel.diag