- Goals next week: Bob & Brian beetle data: what an analysis would be with complete stochastic description.
- Compare models with different
Full model: Parameter list from current code:
/* Biological Parameters. eggs and pupa can get canibalized */
double b = 5; /* Birth rate (per day) */
double u_egg = 0, u_larva = 0.001, u_pupa = 0, u_adult = 0.003; /* Mortality rate (per day) */
double a_egg = 3.8, a_larva = 3.8+16.4, a_pupa = 3.8+16.4+5.0; /* age at which each stage matures, in days */
double cannibal_larva_eggs = 0.01, cannibal_adults_pupa = 0.004, cannibal_adults_eggs = 0.01; /* cannibalism of x on y, per day */
double a_larva_asym = 3.8+8.0; /* age after which larval size asymptotes */- Add individual heterogeneity in egg and larva maturation age and in cannibalism of larva on eggs.
Outline / manuscript draft
Likelihood inference for time series
- All based on step-ahead predictions, from Markov property. Compare to deterministic skeleton’s minimization of sum of squares on step-ahead predictions (i.e. assumes normal deviates).
See the monographs:
- Iacus, S. M. (2008). Simulation and Inference for Stochastic Differential Equations With R Examples. New York: Springer.
- Prakasa Rao, B.L.S. (1999) Statistical Inferences for Diffusion Type Processes, Oxford University Press, New York.
Exploring existing implementations of likelihood methods on SDEs through the R sde package accompanying the Iacus text.
- Many nice methods for SDEs, general case is harder.
Conditions
- Large sample scheme Time interval gets longer with n, while Δ is fixed time-step. Requires the additional assumptions of stationarity and/or ergodicity
- High-frequency scheme: Δ shrinks as n increases, fixed window, need not assume ergodic.
- Rapidly increasing design: hybrid combination with prescribed rate of mesh increase k
Underlying model
dXt = b(Xt,θ)dt + σ(Xt,θ)*d**W*t
Exact Likelihood conditions
- Linear growth assumption:
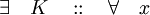
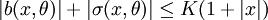
- Global Lipshitz assumption:
b(x,θ) − b(y,θ) | + σ(x,θ) − σ(y,θ) < K | x − y |
Positive diffusion coefficient
Bounded moments
Smooth coefficients (will use up to 3 times differentiable)
Convergence of diffusion part estimator usually 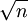 , with
, with 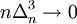
Numerical methods
- Exact likelihood inference (conditional density of process must be known)
- Euler approximation: discretization can assume linearity over small Δt
- Elerian method (Milstein scheme)
- Kessler (higher order Ito/Taylor expansion)
- Simulated likelihood (approximate cdf with subdivisions in timestep over which Euler is accurate).
- Hermite polynomial expansion of likelihood.
#### Steps
- Evaluate the conditional density function
- Evaluate the likelihood function (will be used as single step predictor)
- maximum likelihood estimation
Code updates
- Swapped out my original linked list library for a more intelligent one. Not sure why pointer pointers are so useful but valgrind is happy.
- beetle simulator now creates step-ahead realizations.
- Considerations: replicates in C or R? would be better if R preserved the openmp code, but can always use parallel R to loop over timesteps and benefit from compiled speed on replicates.
- Kernel density estimation for assigning probabilities? Probably reserve at R level at the moment.
Misc Reading & Notes ——————–