Final day of Bodega Phylogenetics workshop: a full day of student presentations. (High resolution images for the trees available by clicking on them)
Principle figures from my investigation of the Labrid data. While morphospace doesn’t present an obvious clustering, it can be effectively divided by my maximum likelihood partitioning scheme on fin angle. A smoothed density plot of fin angle alone reveals two identifiable modes in the traitspace.
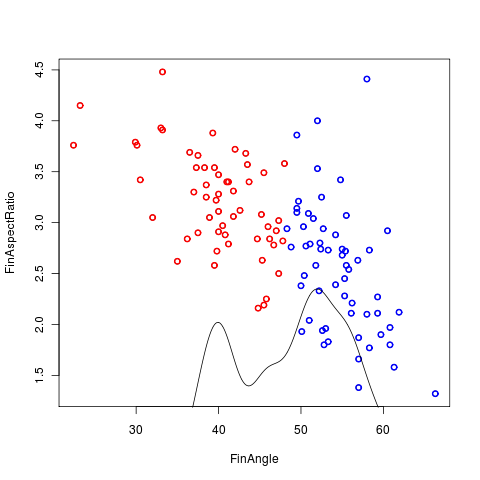
Based on the discretized partition, ancestral states are reconstructed under both the equal rates and independent rates model. Independent rates offers a negligible improvement in likelihood score (-loglik of 72 vs 73)

Without discretizing, I infer ancestral state based on Brownian motion. information is rapidly lost in the inference, and the ancestral states become simple averages of the observed traits.

Using the discrete trait analysis, I assign nodes to their more likely reconstruction and use this as the painting scheme for the inference under continuous traits. A stochastic mapping reconstruction could assign nodes randomly based on the probability inferred by the reconstruction. The full simmap approach could also be taken. I doubt the results would differ much between the approaches, as most of the information is in the tips anyway.
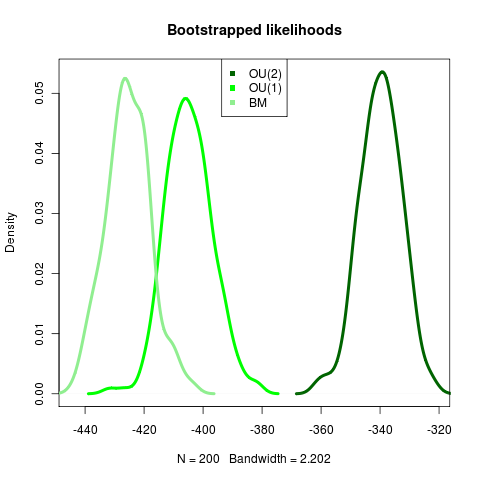
The likelihood distributions generated by bootstrapping 200 replicates shows strong confirmation of the more complex model. Very satisfying in light of the challenges in the Anoles data!
Feedback from the results has been quite compelling and am beginning to write this up a small manuscript. We’ll need to reduce to two figures.