Model choice plays an increasingly dominant role in comparative phylogenetics. The software OUCH of Butler and King (2004) is built around the model comparison framework and returns a variety of information criteria for choosing between different models fitted to the tree.
I’ve written a short R script to explore how robust these inferences are, and the results are rather surprising. None of the inferences holds up to the classical P value standard of 95% probability of being right, and the inference can be wrong more often than it is right – a coin flip out-performs the model choice method based on these information criteria in certain cases. This provides good motivation for my proposed new methods and should also provide material for a note to Evolution.
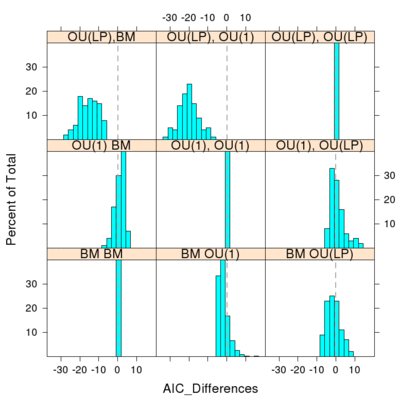
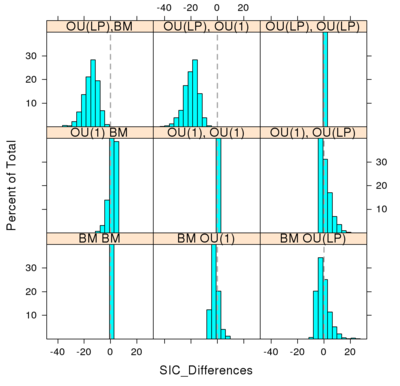
Histogram in the difference between AIC (or SIC) score of the model used to produce the data and the alternative model. Values falling to the left of the dotted line choose the model actually used to create the simulated data on the tree. In the first row, the data is created under the OU(LP) model and the model choice always selects OU(LP), without error. But when the data is created under BM (last row) the OU(1) and OU(LP) models often outperform the BM model. When the data is created under OU(1), middle row, the model inference chooses the other models most of the time. While stricter criteria of SIC make fewer errors relative to AIC when the data is created under the simpler BM model, SIC performs even worse when OU(1) generates the data.
Uses the Anoles tree of the Northern Lesser Antilles and trait of log body size as in the example in OUCH and in Bulter and King paper.
Challenges with Figures
- The figure doesn’t appear exactly as I’d like it to, still mastering R lattice graphics. It fills the rows from left to right but from bottom to top. Also, I’d like to leave off the diagonal (self-comparisons, here appearing as the cross diagonal), since differences between scores is of course zero for these cases. They could be replaced with the labels for those row/columns (BM, OU(1), OU(LP) ).
Code Notes
- Code is implemented in the robust_model_choice.R file in codebranches directory. Uses sapply instead of for loop, but the evaluation of the true and test models for each of the simulated data sets is quite slow still. Not sure if it can be accelerated using OUCH, will try in my own C code implementation. I’ll need to extend the C code to handle OUCH-style painting for this.