Some very good feedback from Hilmar as I tackle some of the semantic capabilities of NeXML in RNeXML. As the complete discussion is already archived in the Github issues tracking, (see in particular #26 and #24) I only paraphrase here.
One of the central advantages we can offer in programmatic generation of NeXML in the R environment is the ability to validate names and enhance the metadata included in the resulting nexml file using programmatic queries to taxonomic name resolution services such as ITIS, EOL, NCBI, as provided in the taxize package.
A subtly to this approach is discussed in issue #24. Whenever we provide new data, we should also provide future users the appropriate metadata describing where it came from, if we had found an exact match or maybe only a close match (perhaps an alternative spelling of the species name was used). Specifying the provenance exactly can become quite verbose, for each taxonomic unit we have:
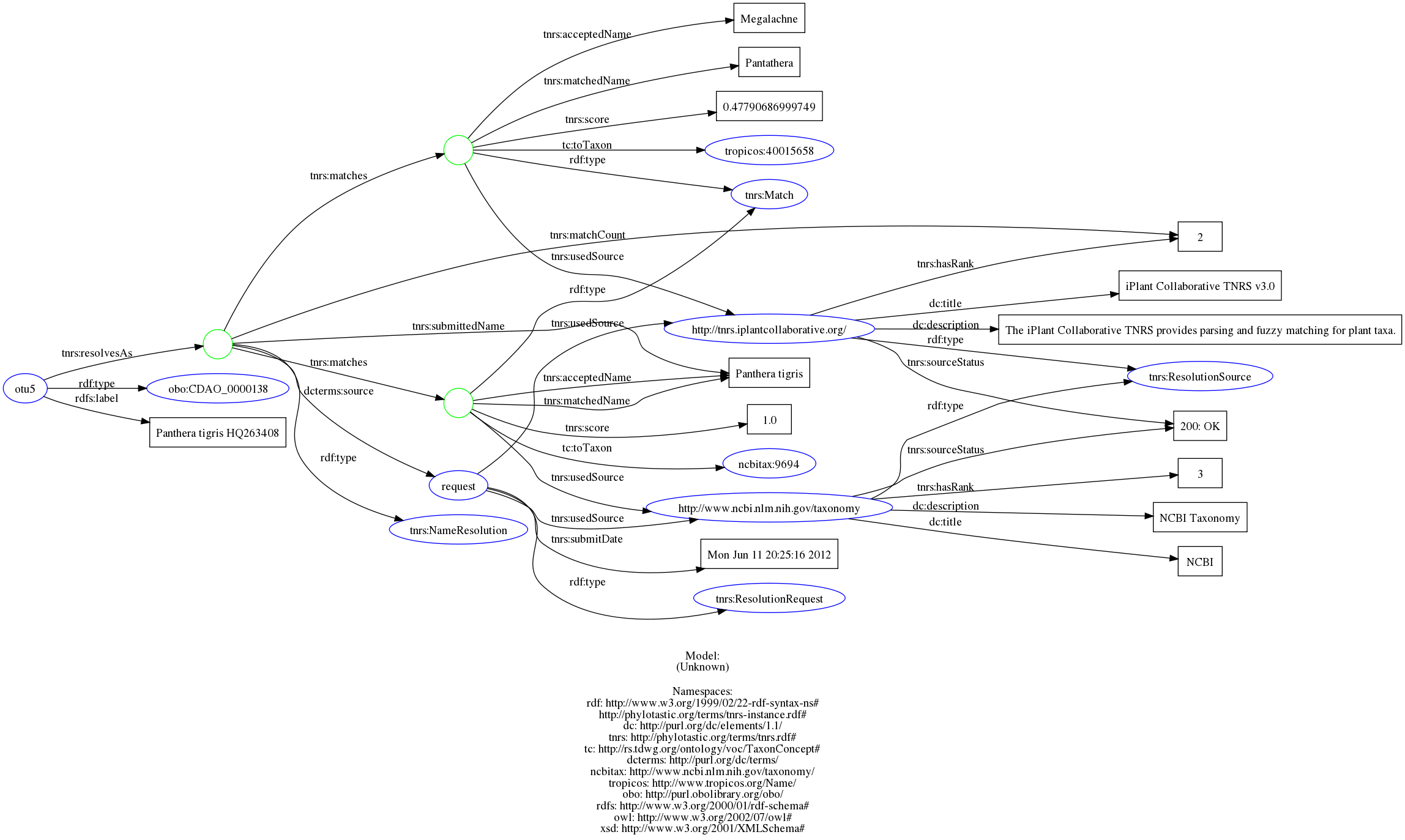
which corresponds to adding RDFa to the NeXML looking something like:
<otus id="tax1">
<otu label="Struthioniformes" id="t1">
<meta xmlns:rdf="https://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:rdfs="https://www.w3.org/2000/01/rdf-schema#"
xmlns="https://www.w3.org/1999/xhtml"
xmlns:obo="https://purl.obolibrary.org/obo/"
xmlns:tc="https://rs.tdwg.org/ontology/voc/TaxonConcept#"
xmlns:dc="https://purl.org/dc/elements/1.1/"
xmlns:dcterms="https://purl.org/dc/terms/"
xmlns:tnrs="https://phylotastic.org/terms/tnrs.rdf#"
xmlns:xsd="https://www.w3.org/2001/XMLSchema#"
class="rdf2rdfa">
<meta class="description" about="https://phylotastic.org/terms/tnrs-instance.rdf#otu5"
typeof="obo:CDAO_0000138">
<meta property="rdfs:label" content="Panthera tigris HQ263408"/>
<meta rel="tnrs:resolvesAs">
<meta class="description" typeof="tnrs:NameResolution">
<meta property="tnrs:matchCount" content="2"/>
<meta rel="tnrs:matches">
<meta class="description" typeof="tnrs:Match">
<meta property="tnrs:acceptedName" content="Panthera tigris"/>
<meta property="tnrs:matchedName" content="Panthera tigris"/>
<meta property="tnrs:score" content="1.0"/>
<meta rel="tc:toTaxon" resource="https://www.ncbi.nlm.nih.gov/taxonomy/9694"/>
<meta rel="tnrs:usedSource">
<meta class="description" about="https://www.ncbi.nlm.nih.gov/taxonomy"
typeof="tnrs:ResolutionSource">
<meta property="dc:description" content="NCBI Taxonomy"/>
<meta property="tnrs:hasRank" content="3"/>
<meta property="tnrs:sourceStatus" content="200: OK"/>
<meta property="dc:title" content="NCBI"/>
</meta>
</meta>
</meta>
</meta>
<meta rel="tnrs:matches">
<meta class="description" typeof="tnrs:Match">
<meta property="tnrs:acceptedName" content="Megalachne"/>
<meta property="tnrs:matchedName" content="Pantathera"/>
<meta property="tnrs:score" content="0.47790686999749"/>
<meta rel="tc:toTaxon" resource="https://www.tropicos.org/Name/40015658"/>
<meta rel="tnrs:usedSource">
<meta class="description" about="https://tnrs.iplantcollaborative.org/"
typeof="tnrs:ResolutionSource">
<meta property="dc:description"
content="The iPlant Collaborative TNRS provides parsing and fuzzy matching for plant taxa."/>
<meta property="tnrs:hasRank" content="2"/>
<meta property="tnrs:sourceStatus" content="200: OK"/>
<meta property="dc:title" content="iPlant Collaborative TNRS v3.0"/>
</meta>
</meta>
</meta>
</meta>
<meta rel="dcterms:source">
<meta class="description"
about="https://phylotastic.org/terms/tnrs-instance.rdf#request"
typeof="tnrs:ResolutionRequest">
<meta property="tnrs:submitDate" content="Mon Jun 11 20:25:16 2012"/>
<meta rel="tnrs:usedSource" resource="https://tnrs.iplantcollaborative.org/"/>
<meta rel="tnrs:usedSource" resource="https://www.ncbi.nlm.nih.gov/taxonomy"/>
</meta>
</meta>
<meta property="tnrs:submittedName" content="Panthera tigris"/>
</meta>
</meta>
</meta>
</meta>
</otu>Why provenance? This can be particularly important in tracking down errors or inconsistencies. For instance, image the taxanomic barcode id number we provide for a taxon is later divided into multiple species. Ideally this would be reflected in the updated entries of the barcode service, establishing new id numbers for the split members and identifying that the old id was split – after all, a barcode system is supposed to facilitate addressing these kinds of issues.
Still, this appears much more verbose than say, what TreeBase does when automatically adding identifiers as annotations to the NeXML otu nodes.
Meanwhile, in more concrete terms, we seem to have some consensus on using NCBI taxonomic ids, since its API queries are pretty fast:
<otus id="tax1">
<otu label="Struthioniformes" id="t1">
<meta xsi:type="ResourceMeta" href="https://ncbi.nlm.nih.gov/taxonomy/8798" rel="tc:toTaxon"/>
</otu>note this example uses the tc: https://rs.tdwg.org/ontology/voc/TaxonConcept# toTaxon concept to provide a ontological definition of the link as a taxon identier.
NCBI does not do partial matching, so we simply warn when a user’s taxonomic names do not match an NCBI id, giving them a chance to correct them if in error (either manually or automatically using the partial name matching functions in taxize)