- This example uses the direct inversion of the covariance function. I have also been exploring the potentially faster Cholksy decomposition without success. Sometimes the match is very good, other times not, see compare-cholsky.md
- This example also on github as reed-example.md.
- Basic likelihood optimization needs to be checked. See commits estimating
sigma_nand estimatingl. Compare the example below to a different set of fixed hyperparameters here
Example
require(pdgControl)
require(ggplot2)Simulate some training data under a stochastic growth function with standard parameterization,
f <- BevHolt
p <- c(1.5, 0.05)
K <- (p[1] - 1)/p[2]Noise function
z_g <- function(sigma_g) rlnorm(1, 0, sigma_g) #1+(2*runif(1, 0, 1)-1)*sigma_g #Parameter definitions
x_grid = seq(0, 1.5 * K, length = 100)
T <- 40
sigma_g <- 0.1
x <- numeric(T)
x[1] <- 1Simulation
for (t in 1:(T - 1)) x[t + 1] = z_g(sigma_g) * f(x[t], h = 0, p = p)
plot(x)
Predict the function over the target grid
obs <- data.frame(x = x[1:(T - 1)], y = x[2:T])
X <- x_grid
library(nonparametricbayes)
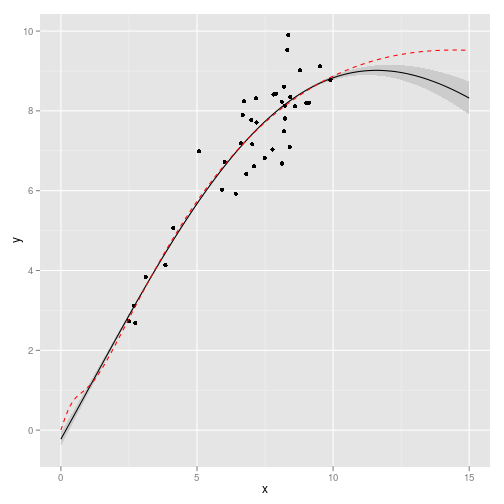
gp <- gp_fit(obs, X, c(sigma_n = 0.05, l = 10))Gaussian Process inference from this model. True model shown in red.
df <- data.frame(x = X, y = gp$Ef, ymin = (gp$Ef - 2 * sqrt(abs(diag(gp$Cf)))),
ymax = (gp$Ef + 2 * sqrt(abs(diag(gp$Cf)))))
true <- data.frame(x = X, y = sapply(X, f, 0, p))
require(ggplot2)
ggplot(df) + geom_ribbon(aes(x, y, ymin = ymin, ymax = ymax), fill = "gray80") +
geom_line(aes(x, y)) + geom_point(data = obs, aes(x, y)) + geom_line(data = true,
aes(x, y), col = "red", lty = 2)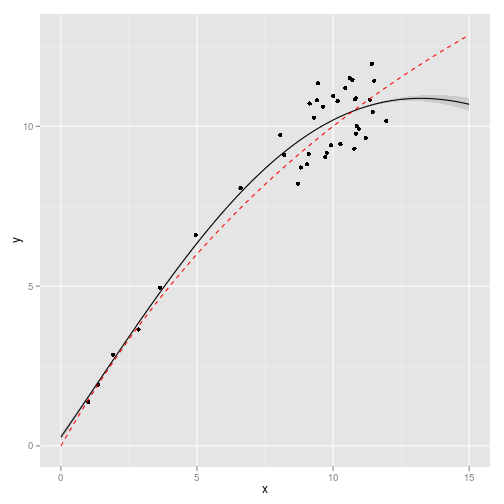
Another example using the May model
f <- May
p <- c(r = 0.75, k = 10, a = 1.2, H = 1, Q = 3)
K <- 8 # approxModel dynamics look like this:
birth <- function(x) p["r"] * (1 - x/p["k"])
death <- function(x) p["a"] * x^(p["Q"] - 1)/(x^p["Q"] + p["H"])
df <- data.frame(x = x_grid, b = sapply(x_grid, birth), d = sapply(x_grid,
death))
ggplot(df) + geom_line(aes(x, b), col = "blue") + geom_line(aes(x,
d), col = "red")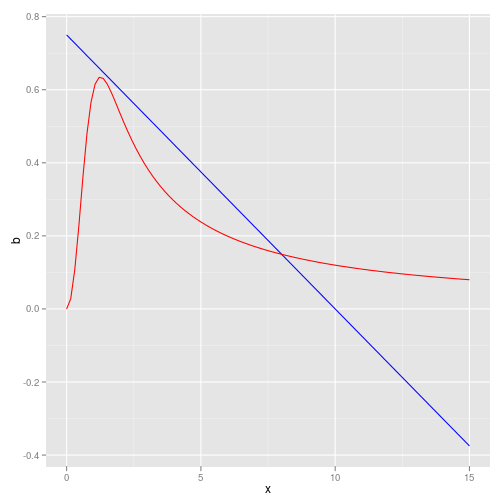
Simulation
x[1] = 2.5
for (t in 1:(T - 1)) x[t + 1] = z_g(sigma_g) * f(x[t], h = 0, p = p)
plot(x)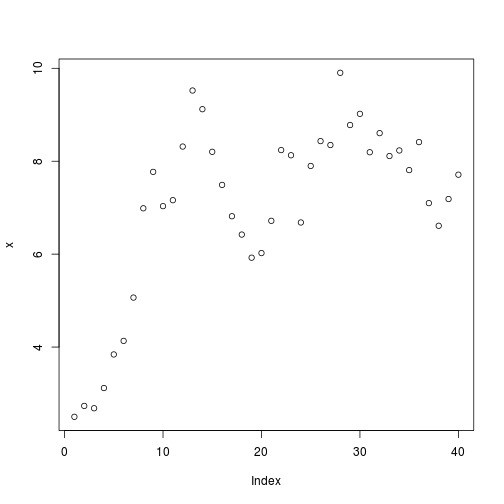
Predict the function over the target grid
obs <- data.frame(x = x[1:(T - 1)], y = x[2:T])
X <- x_grid
library(nonparametricbayes)
gp <- gp_fit(obs, X, c(sigma_n = 0.05, l = 10))Gaussian Process inference from this model
df <- data.frame(x = X, y = gp$Ef, ymin = (gp$Ef - 2 * sqrt(abs(diag(gp$Cf)))),
ymax = (gp$Ef + 2 * sqrt(abs(diag(gp$Cf)))))
true <- data.frame(x = X, y = sapply(X, f, 0, p))
ggplot(df) + geom_ribbon(aes(x, y, ymin = ymin, ymax = ymax), fill = "gray80") +
geom_line(aes(x, y)) + geom_point(data = obs, aes(x, y)) + geom_line(data = true,
aes(x, y), col = "red", lty = 2)