Two-day mini conference with Wainwright lab, Mike Alfaro lab, Rita Mehta lab, Tim Higham lab at the Hastings reserve, with everyone (about 30 of us) presenting, and discussions on convergence and on modularity.
more labrid trait evolution
alpha & sigma models on two shifts, not converged runs. should really consider the mcmc or at least simulated annealing.
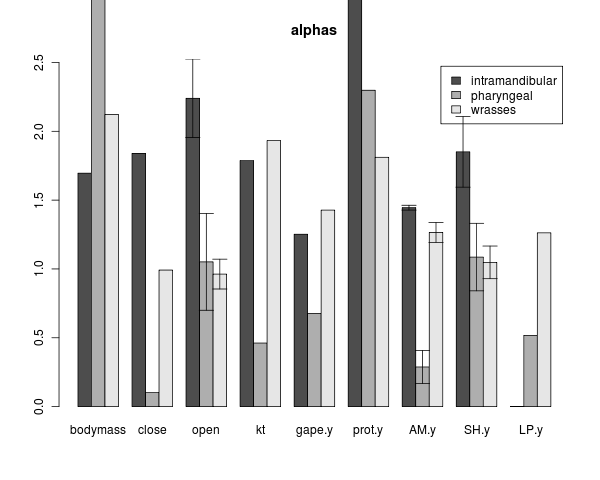
Peter proposes looking at model adequacy by deviance of a observed traits from what is expected under the model (by replicate simulations). Added a little function to do this for my wrightscape fits. Blue: less than 1 sd, green 1-2 sd, orange 2-3, >3 red:

# create a colorcoded plot of how well each tip is fitting it's predicted value
tip_plot <- function(modelfit, n=100, ...){
dat <- modelfit$data
tree <- modelfit$tree
reps <- sapply(1:n, function(i) t(simulate(modelfit)))
m <- rowMeans(reps)
sder <- sapply(1:(dim(reps)[1]), function(i) sd(reps[i,]))
colorcode <- dat[[1]]
colorcode[ abs(dat - m) < 3*sder ] <- "orange"
colorcode[ abs(dat - m) < 2*sder ] <- "green"
colorcode[ abs(dat - m) < sder ] <- "blue"
colorcode[ abs(dat - m) > 3*sder ] <- "red"
tr <- convert(tree, colorcode)
col <- tr$regimes[!is.na(tr$regimes)]
plot(tr, ...)
tiplabels(col=col, pch=19)
list(tree=tr, tipcolors=col)
}Community Phylo exploration
Based on a question from Jonathan Chang’s presentation; we work this out during the break: Evolutionarily-motivated models for null models for community phylogenetics:
picante, distribution of distance-based clustering summary-statistics: mntd, mpd.
Simulate data under the evolutionary transition rate model. Estimate the summary statistic on this data. Bootstrap the estimate over the discrete trait simulations. Not significant:
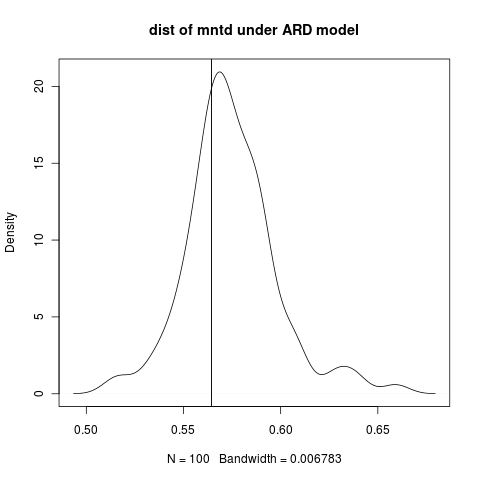
Compare to null distributions from the picante methods (run with ses.mntd, ses.mpd): for instance: p = 0.07 according to ses.mntd on this model.