Working on paper revisions. Added a separate demo file just for generating the figures and analysis used in the manuscript, called likelihood_manuscript.R Need to clean and reorganize the package files and demos.
Added full species names and islands to the tip labels of the tree. Adjusting which example paintings/regimes to include, currently focusing on the original OU.LP model (now calling it OU.3 following Brian’s recommendation for simplicity), OU.4, (originally OU.LP4) and OU.23 (originally all_diff or OU.AD). Paintings below.
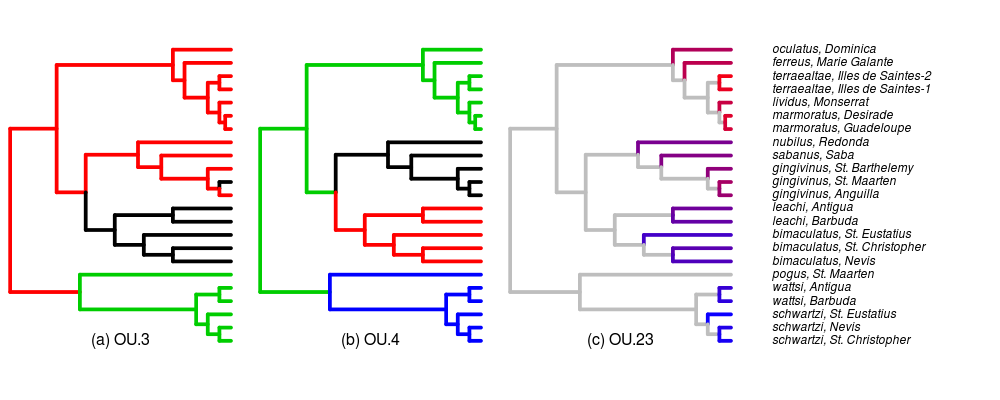
Also queued some more runs of the pairwise comparisons. Modifying the distribution plots to show comparison to AIC and be clearer and more intuitive; more tomorrow.
Diversification Rates
Looking into the extensions of the Monte Carlo likelihood into speciation/extinction rate calculations. Need to first refresh myself on the likelihood calculations for the branching models. Revisiting the literature:
1. Nee, S., May, R.M. & Harvey, P.H. The reconstructed evolutionary process. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 344, 305-311(1994). 2. Nee, S. et al. Extinction rates can be estimated from molecular phylogenies. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 344, 77-82(1994). 3. Kendall, D.G. On the Generalized “Birth-and-Death” Process. Annals of Mathematical Statistics 19, 1-15(1948).
and some of the gieger/laser packages.
Sundry Logistics
Wainwright discussion group – see paper and Friday’s entry.
GTC Guest speaker Scott Dawson on his own research path and “teaching biologists to teach.”
Note to self: I should really hit all my commits to github; flickr etc and hit the publish post on the day I do the work and not the morning after; so as not to confuse myself with the timestamps!