Read through the article for Monday’s PBG270, a 2009 Nature paperfrom the Plotkin lab on mutational robustness and the rate of evolution. They consider an interesting little setup. Start with a Moran model (individual born, may mutate, choses who to replace at random), but allow the following mutational structure (landscape). Of P possible genotypes, each can only move to one of K neighbors by a single mutation. Call the genotype(s) of optimal fitness C. Call all those genotypes that have C as a neighbor (can reach in one mutation) B. All other genotypes that have the same phenotype as B but different K neighbors genotype A. q is the probability that the mutation changes the genotype but not the phenotype; a measure of robustness.
I sketch out the possible outcomes described in the paper, to get a sense of why intermediate values of K/P are optimal.
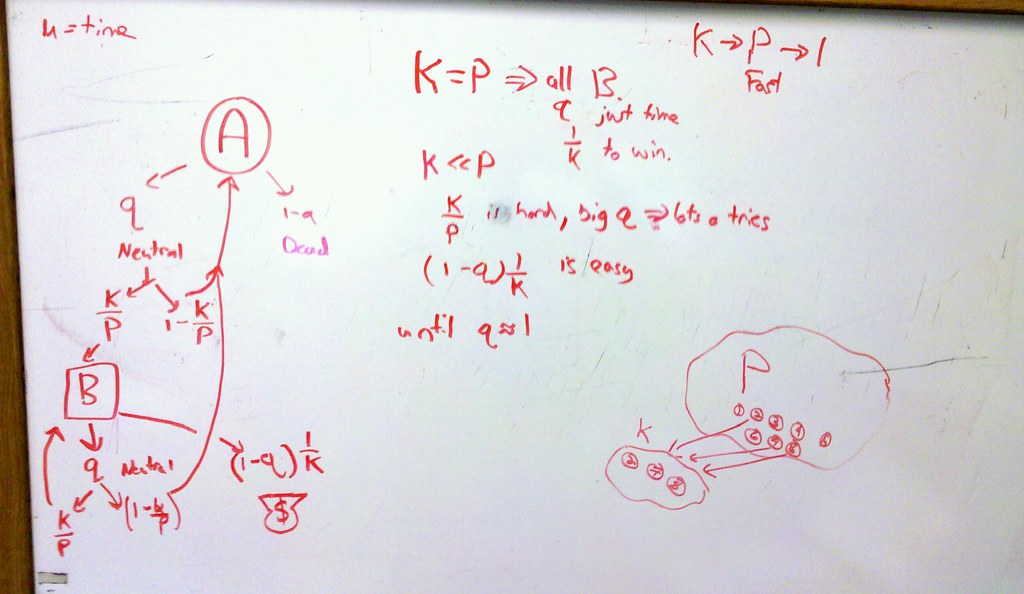
Some things just need a white board. Group discussion on paper Monday.
Phylogenetics Reading
Our Friday Phylogenetics discussion group is converging more and more with Wednesday Monte Carlo with Bruce Rannala. Last week Weds we discussed with Rannala we focused on species tree with the example from BEST: Liu, L. et al. Coalescent methods for estimating phylogenetic trees. Molecular phylogenetics and evolution 53, 320-8(2009).
And that Friday in PDG discussed their eariler paper, where BEST relied on running Mr Bayes separately first, Edwards, S.V., Liu, L. & Pearl, D.K. High-resolution species trees without concatenation. Proceedings of the National Academy of Sciences of the United States of America 104, 5936-41(2007).
This week we moved in Rannala Monte Carlo we ventured into horizontal gene transfer with Linz, S., Radtke, A. & Haeseler, A. von A likelihood framework to measure horizontal gene transfer. Molecular biology and evolution 24, 1312-9(2007).
and continued in PDG along the species-tree literature with the improved performance of star-beast: Heled, J. & Drummond, A.J. B_ayesian inference of species trees from multilocus data. _Molecular biology and evolution 27, 570-80(2010).
Monte Carlo will continue with the BUCKy paper
1. Ané, C. et al. Bayesian estimation of concordance among gene trees. Molecular biology and evolution 24, 412-26(2007).
Seems the software announcement has paper has only just come out in PLoS Comp. Bio this week? :
- Larget, B.R. et al. BUCKy: Gene tree/species tree reconciliation with Bayesian concordance analysis. PLoS computational biology 26, 2910-2911(2010).
In the Wainwright lunch today we also touched upon the recent Sys Biol papers, primarily in reference to diversification rates, 1. Rabosky, D.L. Primary Controls on Species Richness in Higher Taxa. Systematic biology 59, 634-645(2010). 2. Rabosky, D.L. & Alfaro, M.E. Evolutionary Bangs and Whimpers: Methodological Advances and Conceptual Frameworks for Studying Exceptional Diversification. Systematic biology 59, 615-618(2010). 3. Fitzjohn, R.G. Quantitative Traits and Diversification. Systematic biology 59, 619-633(2010).
Most of my notes on the articles are filed with the actual pdfs through Mendeley. Writing here gives me a freedom to interweave how the narrative behind how the papers find my way into my library though. Still figuring out documenting workflow around literature and talks.
Too many meetings, not enough writing…