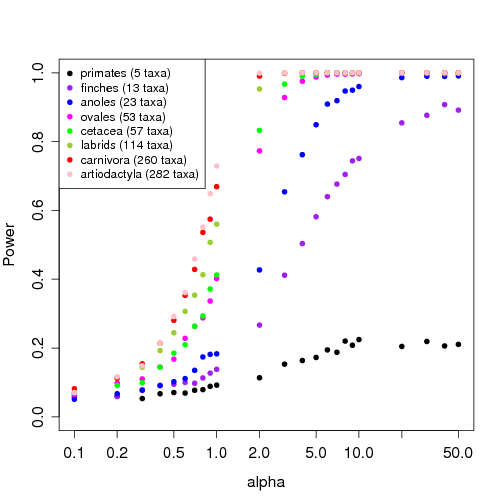
- Inclusion of power figure, discussion of power. (include comparison to smaller trees?)
Code ToDo
Fix parallelization – lower-level parallelization in LR_boot function seems to have high memory and processing overhead. Parallelize over alpha only. Funny that sfSapply at lower level conflicts when inside an upper-level sfSapply call? Can the lower level be put into sequential mode? Requires more experimenting, probably with simple test functions.
Create package around power analysis, including ability to test bootstraps
- Testing of optimizer convergence / error handling in the traitmodels method
- remove traitmodels dependence from power analysis library?
Incorporate faster linear method from traitmodels (at least for bm and ou1) into the wrightscape package…
Power test results ——————
An estimate of how informative a phylogeny is for applications in comparative methods. The independent of trait data, depending only on the structure and size of the tree. (i.e. more taxa help, but unresolved polytomes help less – a large star tree has zero power). All trees are normalized to unit length for comparison of the weakest strength of selection detectable on the tree. For instance, in the 5-taxa primate tree, even if the strength of selection were strong enough to decrease the correlation between ancestors (by e^-2) along a branch only 1/50th of the whole tree length, one would still have only about a 20% chance of detecting that force (at the 95% confidence level). Meanwhile on the carnivora tree one would have about 70% chance of detecting a force of selection 50 times weaker than that, a force that would take the entire time in the tree to decrease the correlation between ancestors by e^-2.