[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2012-01-03 7:00:37” max_upload_date=“2012-01-03 22:23:37”]
kt and open seem the best focal traits. Unclear what good null traits to use would be.
Unclear if anything is gained by indep thetas in this example.
Performance of alpha v theta?
Edited plotting of likelihood comparisons, just grouped by trait to get common axis. (Difficult to get intelligent zooming on facet_grid to ignore the outliers). Trying such as:
lims <- cast(dat, comparison ~ trait, function(x) quantile(x, c(.05,.95)))
coord_cartesian(ylim=c(min(lims), max(lims)), wise=TRUE)but I need different ylims for each row when facet_grid has scales=“free_y”.
[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2012-01-04 7:00:37” max_upload_date=“2012-01-04 13:23:37”]
Running likelihood ratio comparisons across all traits in following model comparisons:
Brownie vs alphas
Brownie vs thetas (ouch/hansen)
thetas vs alphas
sigmas vs alpha-thetas
alphas vs alpha-thetas
thetas vs alpha-thetas
on both two-shifts regime model and the intramandibular regime model.
[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2012-01-06 7:30:37” max_upload_date=“2012-01-06 14:35:00”]
New model comparisons
BM vs Brownie
BM vs OU
Brownie vs alphas
Brownie vs thetas
OU vs Brownie
thetas vs alphas
Labrid data, intramandibular innovation (Summary Plot)
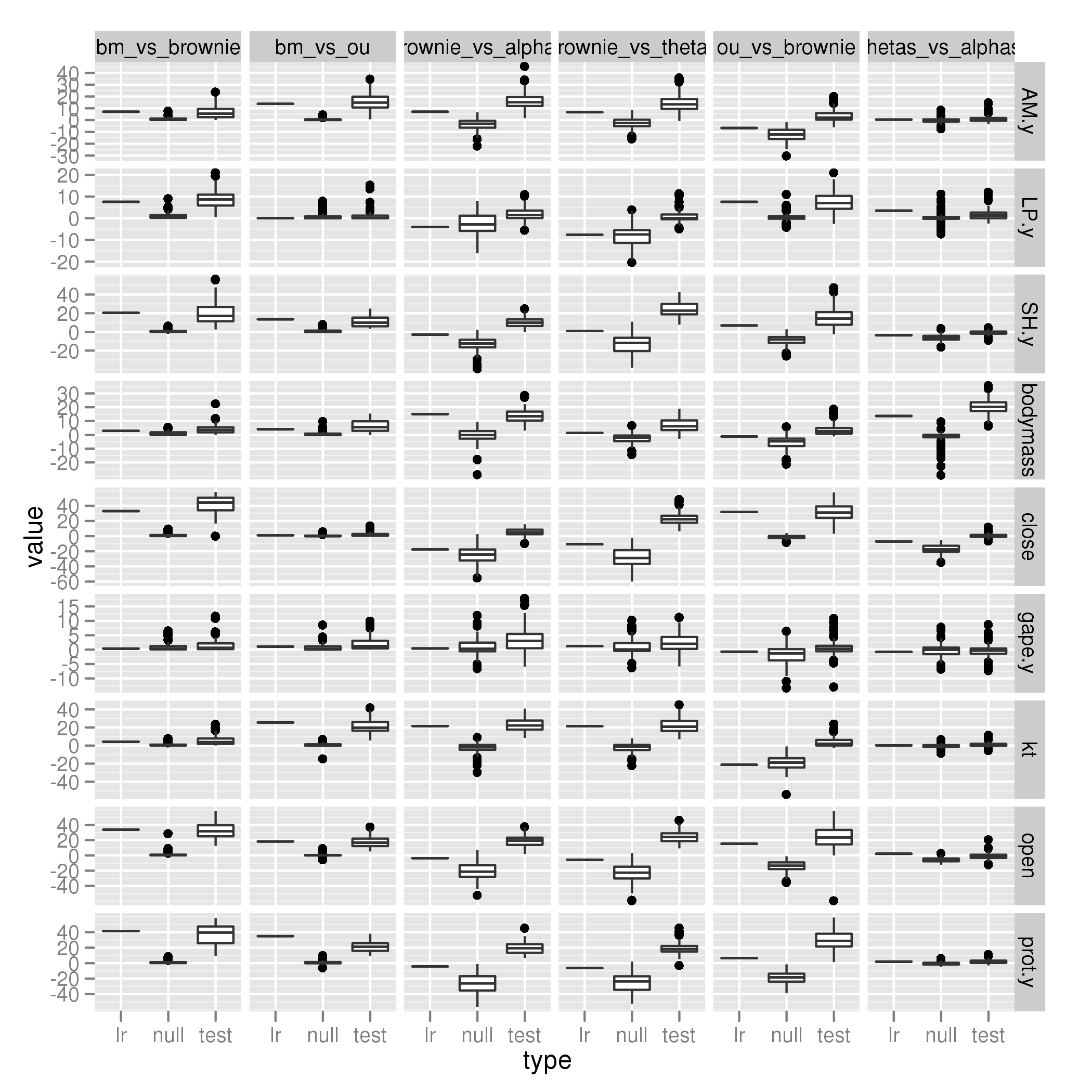
Individual images:
[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2012-01-06 14:35:00” max_upload_date=“2012-01-06 16:00:37”]
Parrotfish only
Not really enough power in Parrotfish-only version:
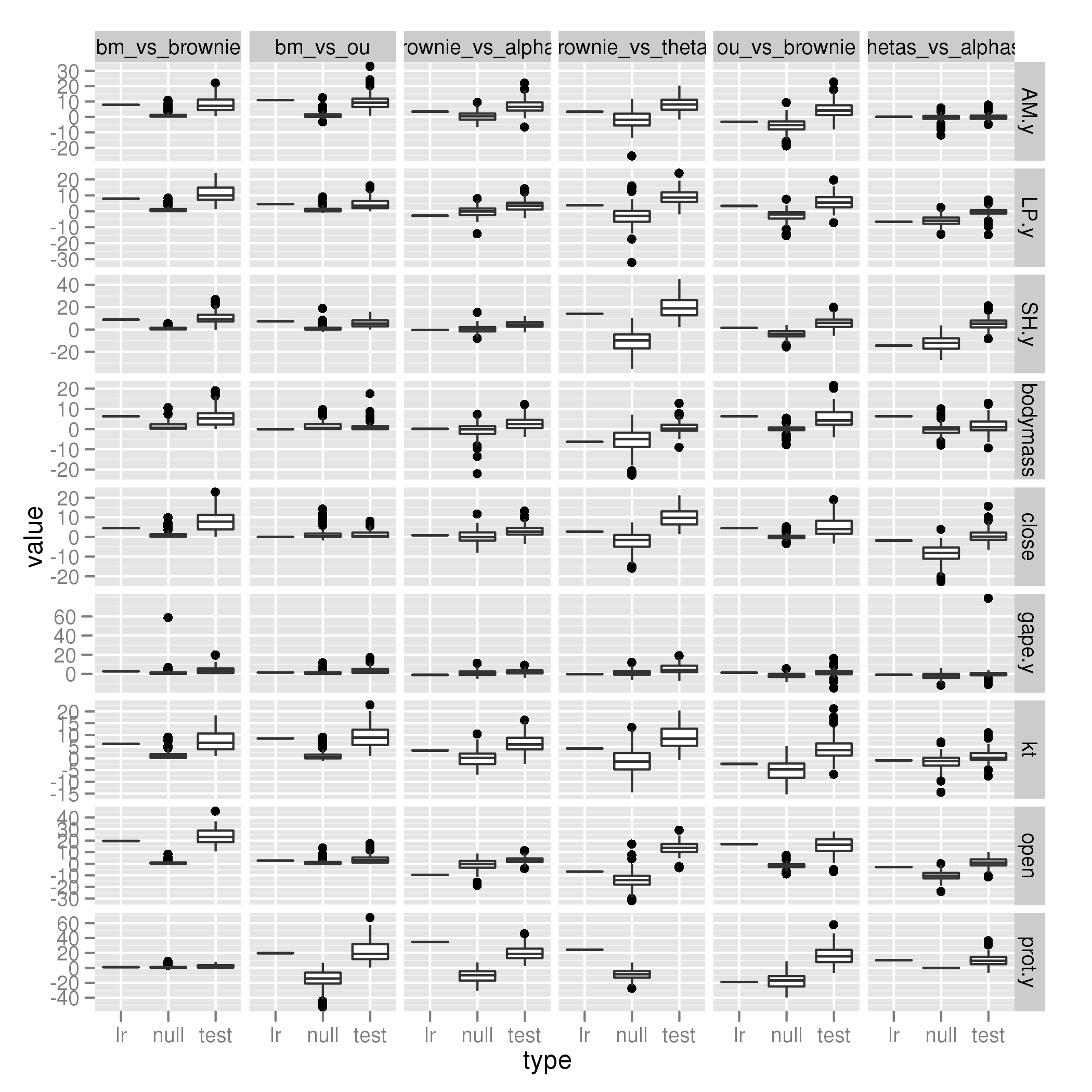
[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2012-01-06 16:05:00” max_upload_date=“2012-01-06 16:30:37”]
Full Labrids
Still running: two-shifts version