Looking at the diversification patterns from Auteur. Longer rjmcmc runs aren’t showing a significant difference, but more attention needed in parameters such as initial merge-split probability perhaps? Seems to overestimate the change points.
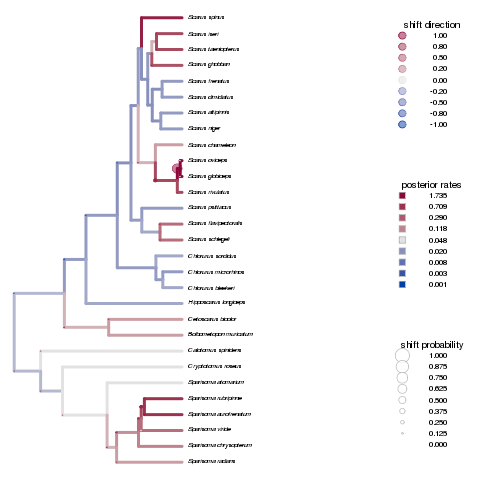
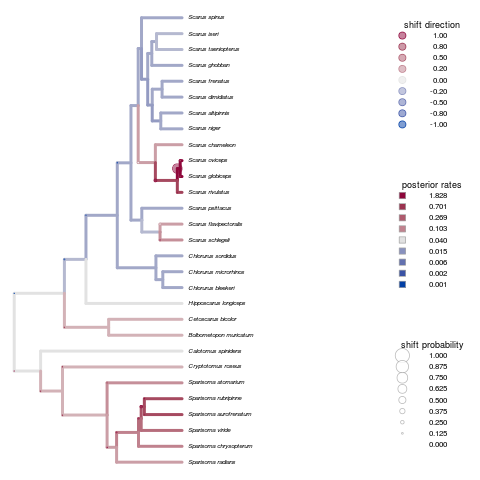
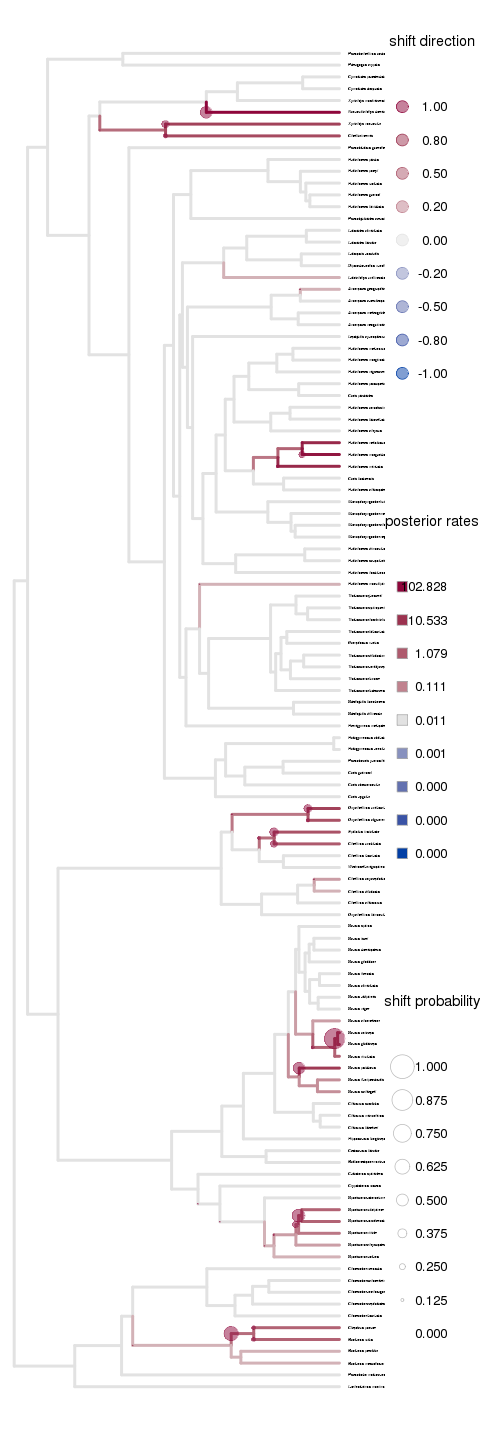
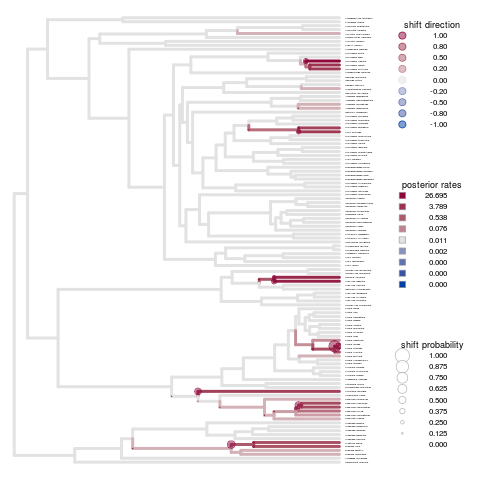
Remains true for the primate brains example:
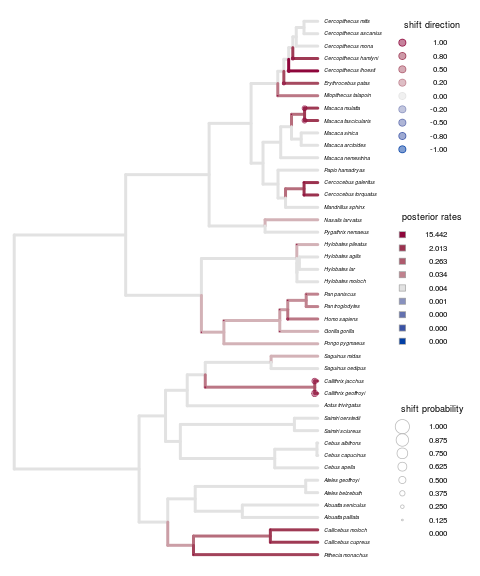
Set up to run on carver cluster, copy over the saved rda file and all files matching the random string already saved as outputs. Full labrid example on 20 cores still exceeds the allocation capacity for an R vector.
Note on the x-axis scale - often isn’t autoscaled, but providing the appropriate a range to xlim (matching max branch length) can fix this.
Cannot figure out how to increase the fontsize of species names - cex or pointsize for png output both only seem to increase fontsize for the legends, etc.