Started writing outline, assembling references
Reviewed brownie-lite as an alternative model to compare the release-of-constraint, but looks like it will be easier just to modify wrightscape code to keep parameters fixed or global.
Writing a wrapper to move the likelihood function for wrightscape to up to the R level.
Figure out which function to pass up: easiest is to build off of fit_model, but just evaluate, don’t optimize the likelihood.
Doxygen Documentation
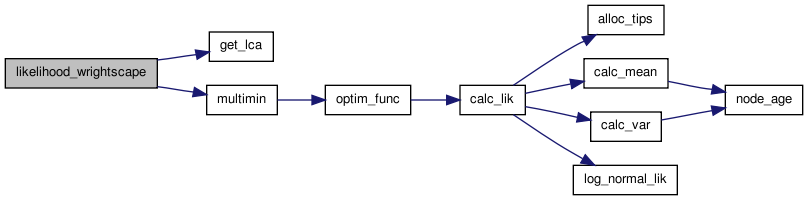
Completing the doxygen documentation of the wrightscape source code to make it easier to navigate
doxygen -g makes the Doxyfile
doxygen Doxyfile makes the documentation
doxygen -u Doxyfile updates the Doxyfile. Not sure when this is necessary
EXTRACT_ALL = YES Extract documentation even from those elements you haven’t yet commented. HAVE_DOT = YES Use Graphviz for class and collaboration diagrammes. CALL_GRAPH = YES Generate a dependency graph for functions and methods. OUTPUT_DIRECTORY = ../doc/Doxygen
-
:Dox
for function documentation,
:DoxLicfor License,
:DoxAuthorfor file header (author, title, etc). See link above for details.
- erg! any typo in latex equation that won’t parse will silently break documentation for rest of file in Doxygen
Code updates
Moving likelihood up to the R level. Steps:
Update function call for calc_lik to conform to the R .C interface (double and integer pointers only). DONE
Write a C wrapper function to call the calculation of the last common ancestor, since we probably don’t want to be repeating this calculation each time. DONE
Write wrapper R function for the calc_lik function DONE
Write example functions calling the optimizer routine on a function defined with a subset of parameters from the full version, to mimic ouch, brownie, etc. DONE
Compare convergence quality and speed