require(MASS)
require(ggplot2)
require(kernlab)Parameterization-specific
X <- seq(-5,5,len=50)
obs <- data.frame(x = c(-4, -3, -1, 0, 2),
y = c(-2, 0, 1, 2, -1))
l <- 1
sigma_n <- 0.8Radial basis function/Gaussian kernel:
SE <- function(Xi,Xj, l) exp(-0.5 * (Xi - Xj) ^ 2 / l ^ 2)
cov <- function(X, Y) outer(X, Y, SE, l) Cholksy method
n <- length(obs$x)
K <- cov(obs$x, obs$x)
I <- diag(1, n)
L <- chol(K + sigma_n^2 * I)
alpha <- solve(t(L), solve(L, obs$y))
k_star <- cov(obs$x, X)
Y <- t(k_star) %*% alpha
v <- solve(L, k_star)
Var <- cov(X,X) - t(v) %*% v
loglik <- -.5 * t(obs$y) %*% alpha - sum(log(diag(L))) - n * log(2 * pi) / 2Direct method
cov_xx_inv <- solve(K + sigma_n^2*I)
Ef <- cov(X, obs$x) %*% cov_xx_inv %*% obs$y
Cf <- cov(X, X) - cov(X, obs$x) %*% cov_xx_inv %*% cov(obs$x, X)gp <- gausspr(obs$x, obs$y, kernel="rbfdot", kpar=list(sigma=1/(2*l^2)), fit=FALSE, scaled=FALSE, var=0.8)
y_p <- predict(gp, X)Things that should be equivelent but aren’t quite:
ggplot(data.frame(x=X, Ef=Ef, Y=Y, y_p = y_p))+ geom_point(aes(x,Ef), col='red') + geom_point(aes(x,Y)) + geom_line(aes(x,y_p))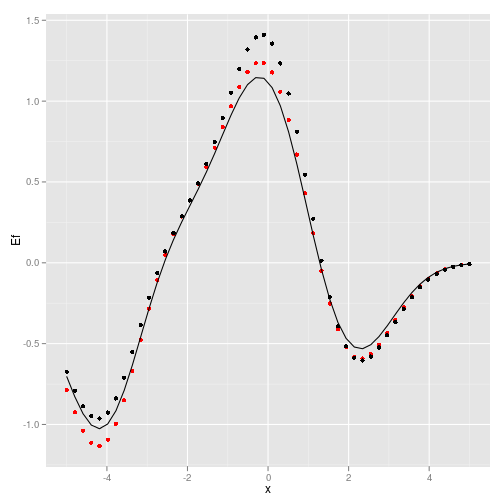
cov_xx_inv %*% obs$y [,1]
[1,] -1.4059
[2,] 0.5010
[3,] 0.1288
[4,] 1.2275
[5,] -0.7119alpha[1] -1.21293 0.46074 0.07548 1.44144 -0.73949alpha(gp) [,1]
[1,] -1.2475
[2,] 0.4011
[3,] 0.1661
[4,] 1.1010
[5,] -0.6394