rfishbase
In answer to Tomomi’s question from last week, have basic functionality as an R package on github, rfishbase. Learned some slightly richer XML parsing in the process.
XML Notes
Had to identify blocks by a child node that specifies the identity, and then find the sibling node that contains the content I needed. Goes something like this:
<dataObject>
<dc:identifier>FB-Size-2</dc:identifier>
<dc:description> Text I need </dc:description>
</dataObject>Which I parsed in R as:
size_node <- getNodeSet(doc, paste("//dc:identifier[string(.) =
'FB-Size-", fish.id, "']/..", sep=""), namespaces)
size <- xmlValue( size_node[[1]][["description"]] )Note that because we specify the exact node, we get only one return item and we can call size_node[[1]]. Note that fish.id is part of the identifier, and the use of the string(.)= to get the content of the identifier. We use xmlValue to get the description element out, though other ways are possible (such as sapply xmlToList). See code for details.
RegExp Notes
Also did some regular expressions parsing for size information, which isn’t intelligently coded in the XML. Found the regexplib website particularly useful for looking up expressions, for instance, for handling numbers with commas and different units. This cheat-sheet also helps.
In particular, getting weight units was a bit tricky:
weight <- gsub(".+published weight: ( (\\d|,)*\\.?\\d*) (g|kg).+", "\\1", size)
weight <- as.numeric(gsub(",", "", weight)) # remove commas
if(units=="kg")
weight <- weight*1000Implementation and Examples
Error handling is quite necessary as there’s lots of missing data. Compute times are a bit slow, but able to create a few smart examples.
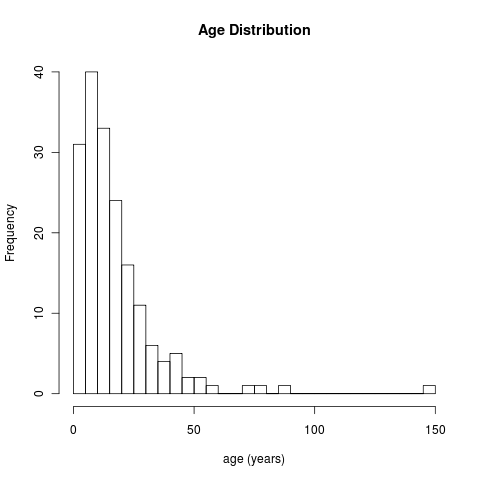
Age distribution for ids 2-500, minus those missing. Image will link to code.
Rmpi
A couple difficulties in my earlier shell scripts on carver/hopper. Re-testing these now. SUCCESS. Test script simple.sh is working on Carver.
simple.sh:
#PBS -l walltime=00:10:00
#PBS -q debug
#PBS -l nodes=2:ppn=8
#PBS -N simple
#PBS -j oe
cd $PBS_O_WORKDIR
module load R/2.12.1
R -f simple.RTest R file:
library("Rmpi")
mpi.spawn.Rslaves(nslaves=15)
slavefn <- function() { print(paste("Hello from", foldNumber)) }
mpi.bcast.cmd(foldNumber <- mpi.comm.rank())
mpi.bcast.Robj2slave(slavefn)
result <- mpi.remote.exec(slavefn())
print(result)
mpi.close.Rslaves()
mpi.quit(save = "no")